Important References
Choose Impact: Excellence and Expertise
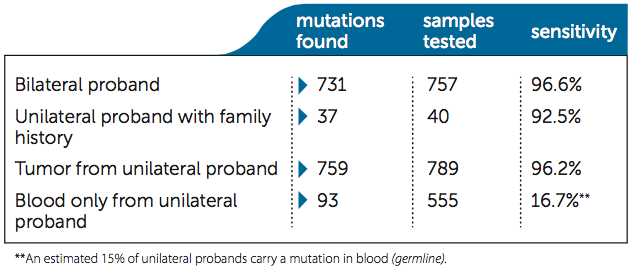
Consider sensitivity when comparing total costs: Higher sensitivity in detecting mutations reduces unnecessary clinical screening for non-carriers. Impact’s diagnostic sensitivity is updated and published in every patient report. As of January 2015, 2141 clinical families have been tested.
MYCN: Impact is the only lab that includes MYCN copy-number testing in retinoblastoma genetic testing.
Reports: We strive to provide as much useful information as possible. World-renowned medical experts contribute to cases involving complex interpretation.
Re-tests: If no mutation is found, we bank any remaining DNA and RNA and re-test in the future when new science or test methods are available. For any new findings, we re-issue our report at no added charge.
Experience: We have completed analysis for more than 2057 retinoblastoma families, building a knowledge base that allows us to provide more accurate interpretation.
Ascribing value to variants: We analyze all variants of uncertain significance using current in silico methods and take into account the knowledge we have gained from our previous test experience. In addition, we search RB1 mutation databases for previous reports of the variant in question. Where needed to define a causative mutation, we request family member samples to test at no added charge.
Clinically appropriate turn-around time: Proband turn around time is 3-6 weeks. Relative turn-around time is 2-3 weeks. Prenatal is 7 working days.
Service excellence: Impact is committed to exceptional customer service. Our team happily provides test order support so you can spend more time with your patients.
Logistics: Impact provides genetic testing services to about 25 countries and samples are routinely shipped from across the world without disruption. We provide the necessary paperwork and recommend a courier service that will reliably deliver samples.
Test Description
Genes tested: RB1, MYCN
Copy number changes: Our lab uses MLPA multiplex ligation dependent probe amplification to look for whole-exon and multi-exon deletions and duplications in the RB1 gene; this method simultaneously screens for small intra-exon insertions and deletions in RB1. Our lab also performs a QM-PCR test to measure MYCN copy number in any tumor sample that does not have an RB1 mutation. This test detects a new sub-set of retinoblastoma with early age at diagnosis, distinct histology, no RB1 mutations, and huge amplification of the MYCN oncogene. [Rushlow, D. et al. 2013. Lancet Oncology.]
Sequence analysis: Our lab sequences the RB1 gene core promoter and exons 1 through 25, as well as nearby flanking intronic regions. Our sequence analysis is able to detect mosaic mutations at a level of 15% or greater. We consider reported polymorphisms when designing our sequencing assays to ensure the accuracy of our sequence results.
Splice site analysis: We sequence a minimum of 25 nucleotides flanking each exon of RB1 to detect changes in splice sites. We use in silico analysis and scoring to determine whether a particular change is likely to cause missplicing. In the case of an intronic variant of uncertain significance we perform RNA transcript analysis on a fresh blood sample at no added charge.
Allele-specific PCR (AS-PCR) for eleven recurrent RB1 mutations: Our lab uses multiplex AS-PCR screens for rapid detection of eleven recurrent RB1 mutations, which are then confirmed by sequence analysis. The highly sensitive AS-PCR can detect the eleven mutations at mosaic levels as low as 1% mutant DNA.
Testing for methylation of the RB1 promoter: Aberrant methylation of the RB1 promoter leads to reduced transcription of RB1, and can initiate unilateral sporadic retinoblastoma in the absence of an RB1 mutation. Our lab identifies RB1 promoter methylation leading to retinoblastoma in approximately 12% of unilateral sporadic tumors.
Amino acid conservation analysis: For missense amino acid changes of uncertain significance we employ several in silico conservation analysis programs to predict whether a particular missense change is likely to be pathogenic. In addition, when appropriate we will test known affected and unaffected relatives at no charge to clarify variant classification.
Further Reference Links
- Impact’s retinoblastoma research articles, including the paper on a new form of retinoblastoma.
- National Cancer Institute Review for Retinoblastoma
- Children’s Cancer Web, Resources for Health Professionals
- US National Library of Medicine Review for Retinoblastoma
- RB1 Gene Mutation Database
- GeneReviews Retinoblastoma Entry